Section 9 SUBNETWORK ANALYSER (GENES) - SAG
9.1 Input Interface
Input
Step 1: Enter a list of genes, with the 1st column for gene symbols and the 2nd column for the significance info (p-values between 0 and 1).Step 2andStep 3: Identify a subnetwork of highly-scored genes from a gene network (Step 2), with the desired number of genes in the resulting subnetwork (Step 3). The significance (p-value) of observing the identified subnetwork by chance can also be estimated by a degree-preserving node permutation test.More Controls: Use additional parameters to fine-tune the steps described above.SUBMIT: click the SUBMIT button to execute the analysis.
Output
- Example Output includes interactive tables and high-quality figures for the resulting subnetwork. A summary of input data and the runtime (computed on the server side) is also provided for reference.
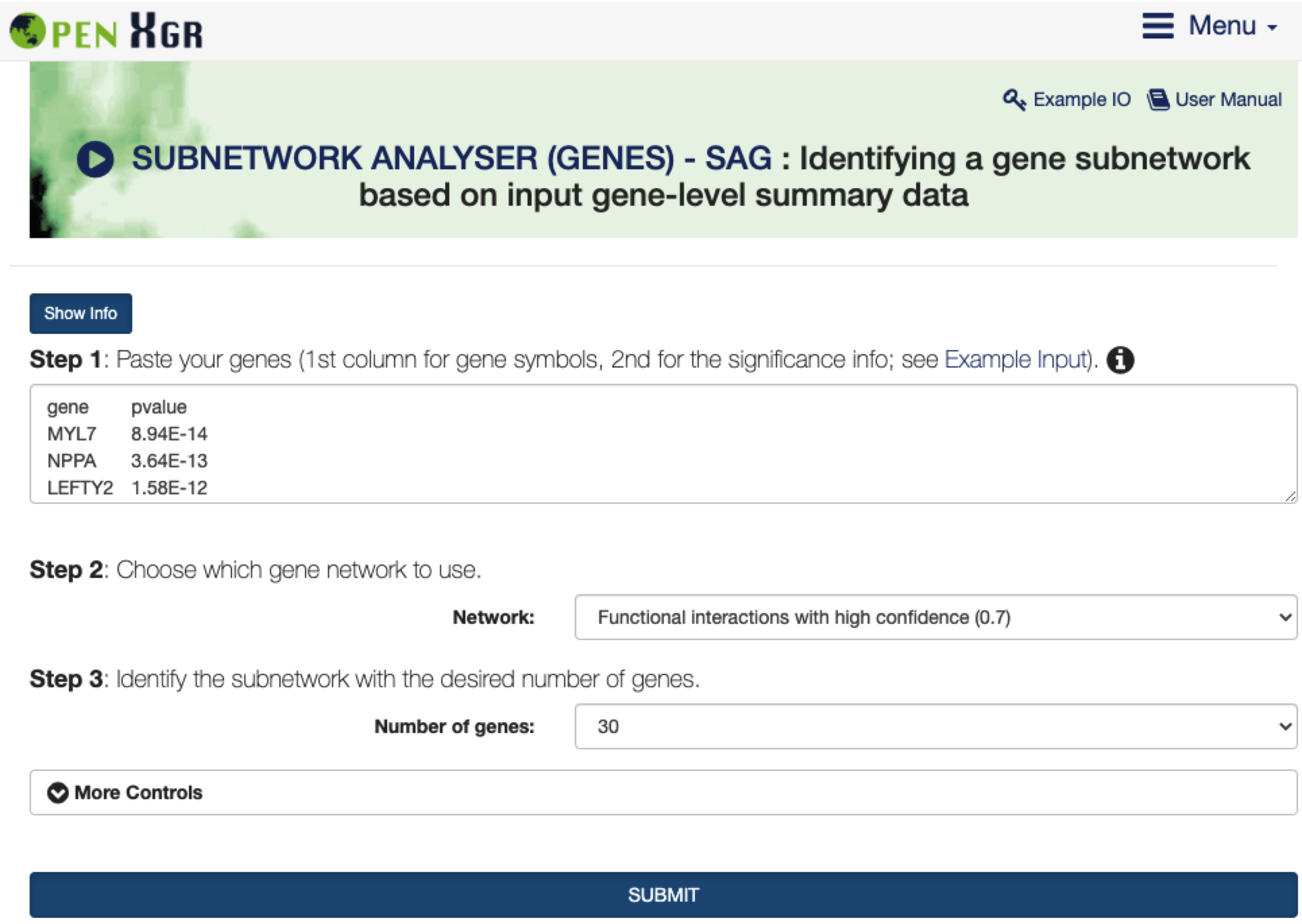
FIGURE 9.1: The interface of SUBNETWORK ANALYSER (GENES) - SAG. The Show/Hide Info toggle button contains the help information on how to use it, including inputs, outputs, and other relevant information.
9.2 Subnetwork Results
- Under the
Input Gene-Level Summary Datatab,An interactive tablelists user-input summary data.
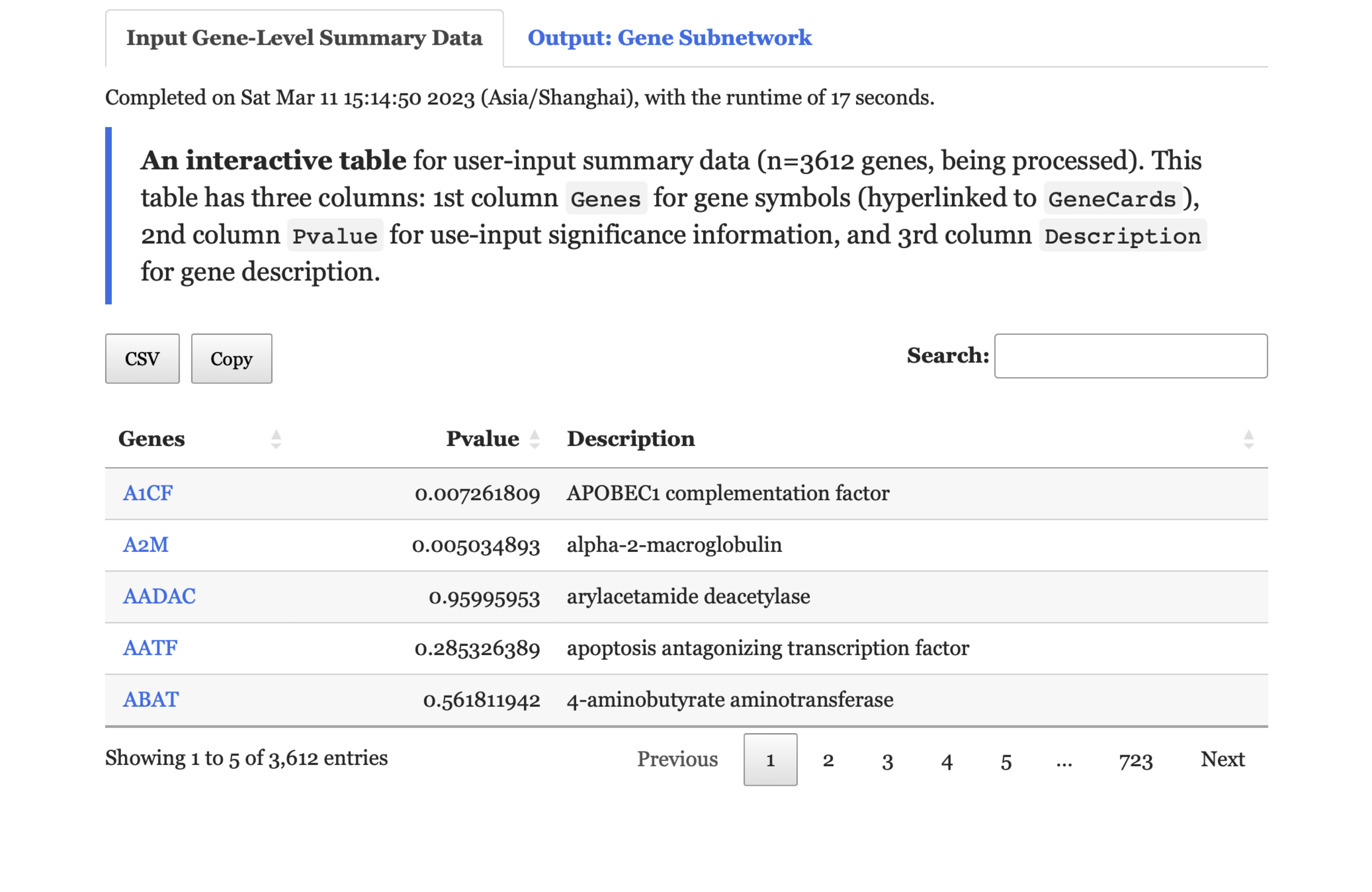
FIGURE 9.2: The user-input summary under the Input Gene-Level Summary Data tab returned for exploration.
- Under the
Output: Gene Subnetworktab,A network visualisationof the subnetwork is provided, along withAn interactive tablefor the subnetwork genes.
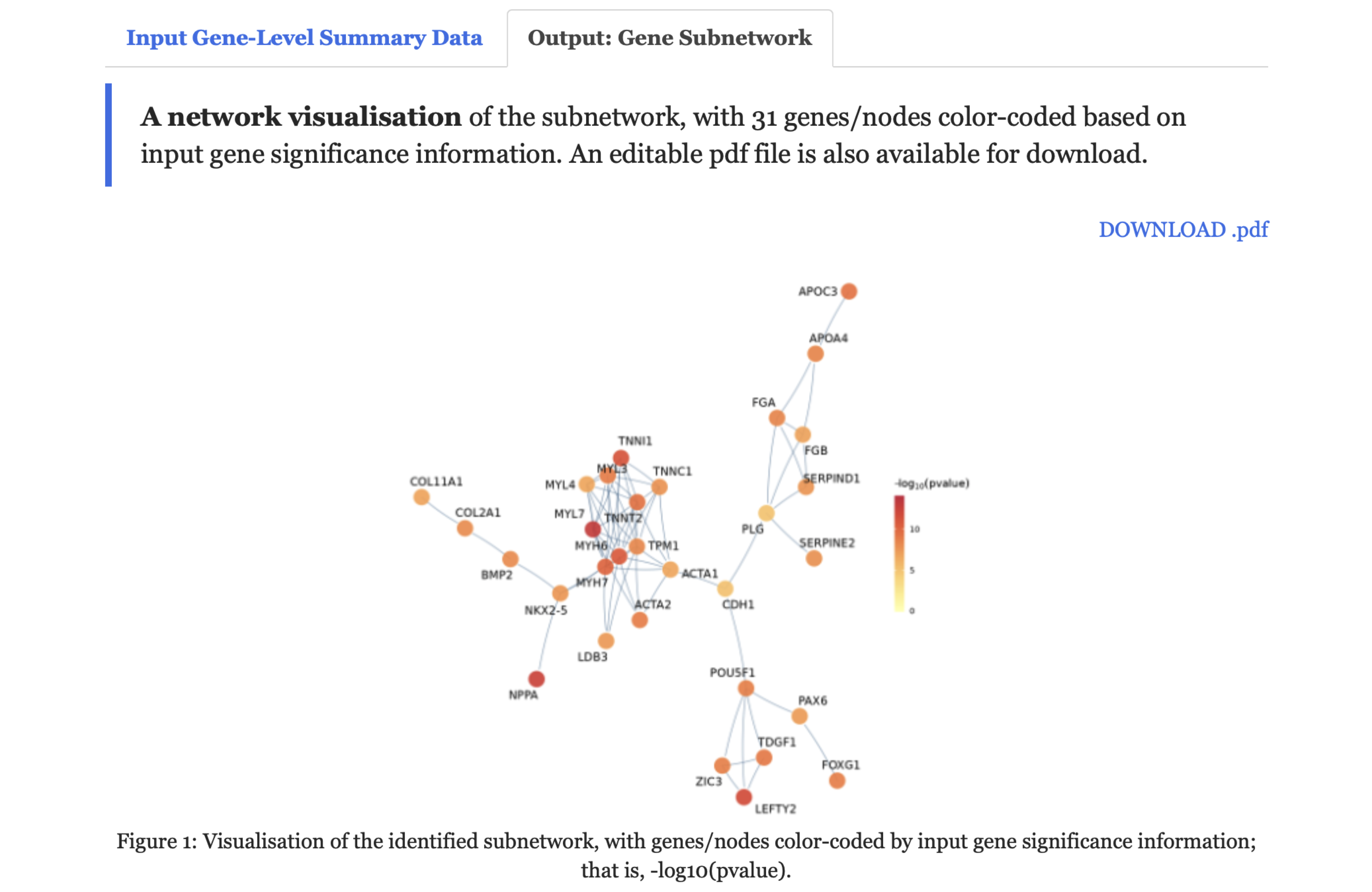
FIGURE 9.3: Visualisation of the identified subnetwork, with genes/nodes color-coded by input gene significance information under the Output: Gene Subnetwork tab.